The pig industry is one the main pillars of livestock production in Japan and domestic pork production ranks 8th in the world. The major focus of research on pig genomics is to develop genomic resources of livestock animals and provide information to contribute in the development of high-quality, safe and secure livestock products. Research on pig genomics was initiated through the Animal Genome Research Project, in collaboration with the Institute of the Society for Techno-innovation of Agriculture, Forestry and Fisheries (STAFF-Institute) and other related research institutes. The project focused on elucidating the genomic structure and function of livestock. Subsequently, sequencing of the genome has been undertaken in collaboration with the International Swine Genome Sequencing Consortium. Additionally, more than 10,000 expressed genes (full length cDNAs) have been analyzed to facilitate the characterization of annotated genes in the genome. The genome information is used to develop methods for discrimination of breeds or individuals by using DNA polymorphism, identification of the genes responsible for the number of vertebrae and meat color in pigs, and developing methods for detecting the association between gene polymorphisms and phenotype. With the effective utilization of genomic information, the ultimate goal is to identify the genes responsible for meat quality, growth rate, disease resistance, reproductive performance, and other traits, to develop gene markers that support rapid breed improvement, and facilitate the development of a variety of products corresponding to various needs of consumers.
- Pig genome sequencing
- Pig genomics databases and resources
- The pig genome sequence and implications in livestock production
Pig genome sequencing

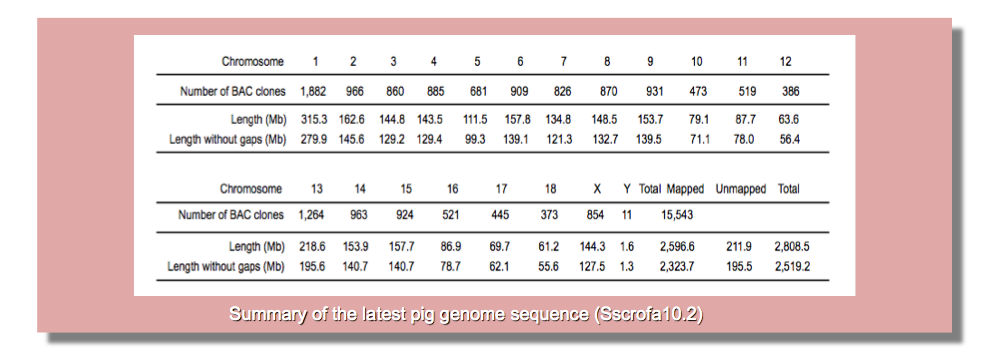 Genome sequencing and isolation of responsible genes are important for effective breeding of pigs for valuable economic traits such as disease resistance, productivity, growth rate, and meat quality. On the other hand, genome sequence and precise information of genes is valuable for utilization of pig as a model animal for biomedical research. In collaboration with the International Swine Genome Sequencing Consortium (ISGSC), we contributed to the sequencing of the entire pig genome. We also precisely sequenced and characterized the genes on the pig genome using full-length cDNA libraries that eventually led to an accurate estimation of the number and structure of genes in the entire genome. We took charge of sequencing of genomic DNA clones located in chromosomes 6 and 7 derived from the Duroc pig (Sus scrofa), an important breed for meat production worldwide. The SGSC high-quality genome sequence corresponds to 90% (2.52 Gb) of the pig genome (approximately 2.8 Gb). Additionally, a collection of 31,079 pig full-length cDNAs generated from various organs and cell populations has been developed. Comparison of the cDNA clone sequences and the pig genome sequence clarified the locations of the full-length cDNAs on the pig genome. The sequenced cDNAs were estimated to correspond to approximately 15,000 genes, and greatly contributed in characterizing the structure of approximately 25,000 genes on the pig genome.
Genome sequencing and isolation of responsible genes are important for effective breeding of pigs for valuable economic traits such as disease resistance, productivity, growth rate, and meat quality. On the other hand, genome sequence and precise information of genes is valuable for utilization of pig as a model animal for biomedical research. In collaboration with the International Swine Genome Sequencing Consortium (ISGSC), we contributed to the sequencing of the entire pig genome. We also precisely sequenced and characterized the genes on the pig genome using full-length cDNA libraries that eventually led to an accurate estimation of the number and structure of genes in the entire genome. We took charge of sequencing of genomic DNA clones located in chromosomes 6 and 7 derived from the Duroc pig (Sus scrofa), an important breed for meat production worldwide. The SGSC high-quality genome sequence corresponds to 90% (2.52 Gb) of the pig genome (approximately 2.8 Gb). Additionally, a collection of 31,079 pig full-length cDNAs generated from various organs and cell populations has been developed. Comparison of the cDNA clone sequences and the pig genome sequence clarified the locations of the full-length cDNAs on the pig genome. The sequenced cDNAs were estimated to correspond to approximately 15,000 genes, and greatly contributed in characterizing the structure of approximately 25,000 genes on the pig genome.
- Groenen M.A.M, Archibald A.L, Uenishi H et al. (2012) Analyses of pig genomes provide insight into porcine demography and evolution Nature 491(7424): 393-398
- Uenishi H, Morozumi T, Toki D, Eguchi-Ogawa T, Rund L.A, Schook L.B (2012) Large-scale sequencing based on full-length-enriched cDNA libraries in pigs: contribution to annotation of the pig genome draft sequence BMC Genomics 13: 581
- International Swine Genome Sequencing Consortium
Pig genomics databases and resources
Pig genomics databases have been developed to facilitate efficient utilization of the genome information in various aspects of pig research. To catalogue the full-length mRNA sequences expressed in pigs, full-length cDNA libraries were constructed from various porcine tissues such as thymus, spleen, ovary, liver, peripheral blood mononuclear cells, uterus, lung, mesenteric lymph nodes and trachea derived from western crossbred pigs and Meishan pigs. A large-scale EST analysis from 5'-end was performed and the resulting 107,068 reads were clustered and assembled into 6,655 contigs and 41,487 singlets. These sequences were subjected to BLAST search against the sequences of mammalian genes (UniGene and RefSeq in NCBI) and the human genome. The assembled sequences and BLAST results were incorporated in a database, PEDE (Pig EST Data Explorer, http://pede.dna.affrc.go.jp/), equipped with web interfaces for various kinds of searches. The PEDE database provides porcine cDNA clones, the inserts of which are estimated to include the full-length sequence of at least 6,000 different genes. Furthermore, PEDE also contains more than 1,200 SNPs extracted in the contigs, which are classified by their origin and effect on the coding amino acids. The SNPs can be utilized for linkage analysis in experimental resource families. The PEDE database will help users to explore genes that may be responsible for traits like disease susceptibility.
Pig genomics databases
Pig genomics resources
The pig genome sequence and implications in livestock production
QTL affecting the number of vertebrae in pigs

The number of vertebrae in swine which is associated with body length varies thereby affecting productivity. The QTL for the number of vertebrae were detected by linkage analyses using F2 experimental families. Detailed analysis using DNA markers developed in the genome sequence of a QTL region on swine chromosome 1 revealed that a 250-kb region is fixed in the pig breeds with a large number of vertebrae.







