NARO has launched GenEditScan, a free and user-friendly analysis tool available on its website. This tool uses the k-mer method to detect foreign DNA in genome-edited crops through next-generation sequencing. With its straightforward design, GenEditScan simplifies the analysis process and generates the essential data needed to confirm the absence of foreign DNA in genome-edited crops. By facilitating such verification, GenEditScan is expected to significantly advance research in genome-edited crops.
Overview
Crop breeding technology uses genetic modification to improve crop characteristics. Traditionally, these advancements have relied on natural mutations or artificial methods, but in recent years there has been a surge of interest in genome editing. One notable technique, deletion-type genome editing, allows precise targeting and cutting of specific DNA sequences, resulting in mutations in target genes. This method has facilitated the efficient development of innovative crop varieties, such as tomatoes with enhanced GABA level.
In the common process of creating genome-edited crops, foreign DNA is temporarily introduced to produce enzymes that cut the target DNA. To classify such crops as genome-edited, it is critical to confirm the complete removal of this foreign DNA. To address this issue, NARO developed the k-mer method, a breakthrough technique that uses next-generation sequencing to analyze vast amounts of DNA sequence data and identify foreign DNA sequences efficiently and accurately through statistical testing. This tool was first made publicly available in 2020.
While the original tool was effective, its complexity and susceptibility to false positives prompted the development of an improved version, GenEditScan. Available at https://github.com/hirsakai/GenEditScan, this enhanced tool streamlines the analysis of genomic data. By incorporating corrected statistical tests, GenEditScan simplifies the process and improves the accuracy of detecting foreign DNA. Validation tests with actual genome-edited crops have confirmed its reliability.
GenEditScan is expected to become a widely used tool that provides researchers with an accessible and accurate method to verify the absence of foreign DNA in genome-edited crops, thereby advancing the field of genome editing.
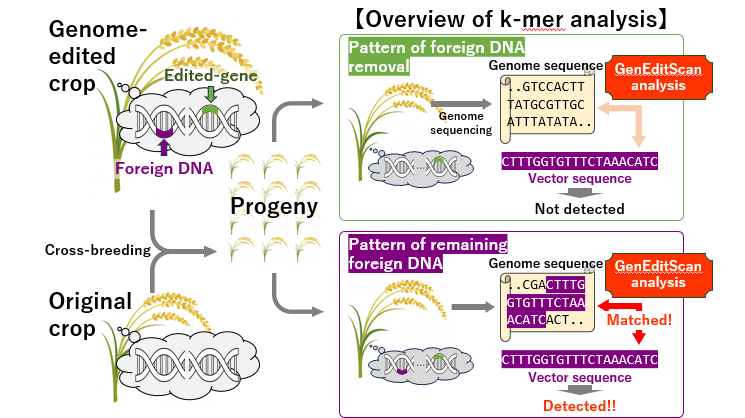
Fig. 1: An example of the process of removing the enzyme genes that remain in the genome-edited individual and an overview of the k-mer method.
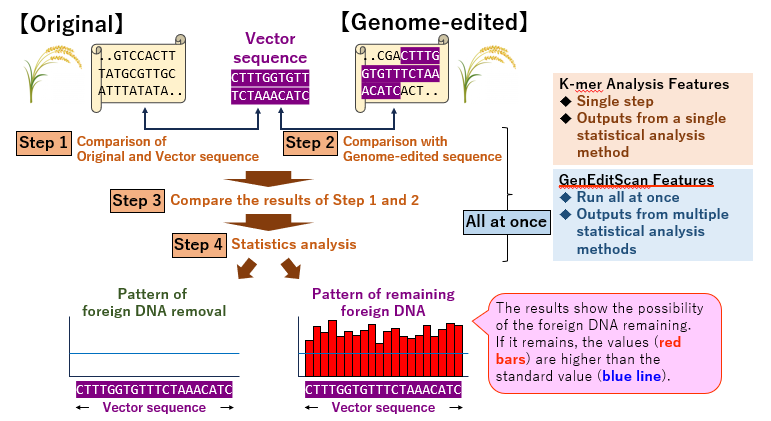
Fig. 2: Flow and features of the k-mer method analysis tool and GenEditScan
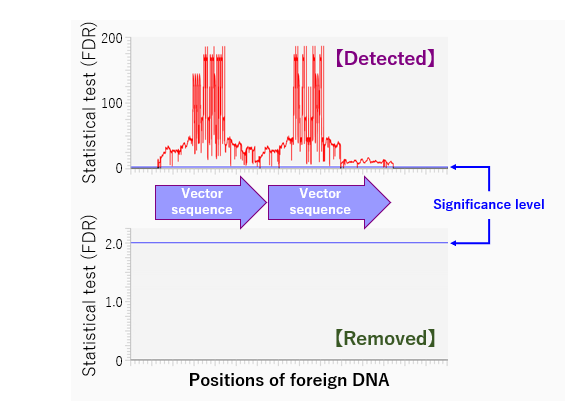
Fig.3: Example of the use of GenEditScan.
Graph generated by analyzing the genome sequence (DDBJ:DRA015756) published in Yasumoto and Muranaka (2023) (https://doi.org/10.1038/s41598-023-38897-x) using GenEditScan. The horizontal axis of the graph shows the location of the foreign DNA and the vertical axis shows the statistical test volume (FDR control) (the blue horizontal line is the 1% significance level line). When foreign DNA remains, a peak is usually detected in the region where the foreign DNA is present (red line), as shown in the figure above. On the other hand, if no genome-editing enzyme genes remain, peaks above the significance level (blue horizontal line) will generally not be detected, as shown in the figure below.
Budget
This work was supported by the MAFF commissioned project study on "Accumulation of scientific knowledge for promoting public understanding of genome editing technologies" Grant Number JPJ008723.




