The genomes of 11 high pathogenicity avian influenza virus in domestic poultry during the early stages of the 2024 season in Japan were analyzed and genotyped. Four distinct genotypes were identified, including one that has been detected for four consecutive seasons and three newly emerging genotypes. As these genotypes were detected within the first six weeks of the season, ongoing vigilance is essential to prevent virus entry into poultry facilities.
Overview
The first outbreak of high pathogenicity avian influenza (HPAI) in domestic poultry for the 2024 season was reported on October 17 last year. By January 22, 2025, a total of 42 cases had been confirmed at poultry feeding facilities.
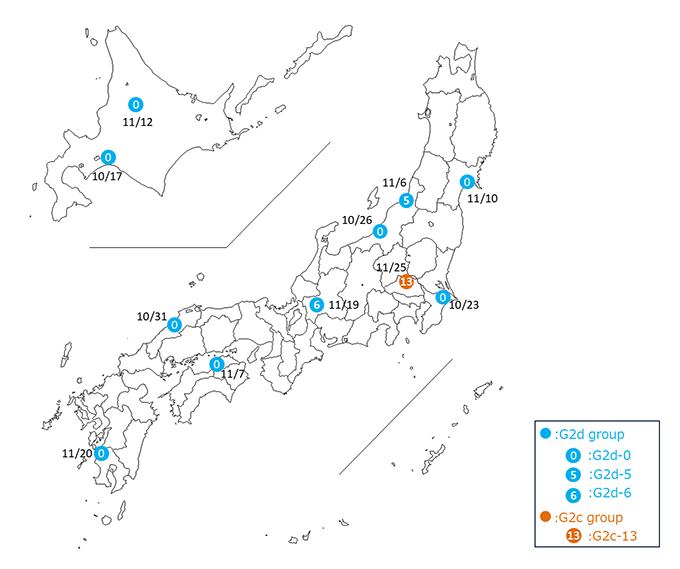
NARO conducted whole genome sequencing on 11 H5N1 HPAI virus strains isolated from outbreaks that occurred within six weeks of the initial case (October-November 2024). The analysis classified the viruses into four distinct genotypes: G2d-0 (8 strains), G2d-5 (1 strain), G2d-6 (1 strain), and G2c-13 (1 strain). Among these, G2d-0 was the most prevalent. Notably, G2d-0 has been consistently detected in Japan for four consecutive seasons since 2021. The same virus type was also identified in wild birds in Hokkaido on September 30, prior to the first poultry outbreak, suggesting that migratory birds introduced the virus during the 2024 season.
The other three genotypes (G2d-5, G2d-6, and G2c-13) were newly identified in 2024. Genetic analysis revealed that some segments of these viruses originated from avian influenza viruses and HPAI strains detected in wild birds both in Japan and abroad. This genetic reassortment likely occurred due to repeated infections among wild bird populations, such as wild ducks (mallards).
Further studies using intranasal inoculation--the natural route of infection in chickens--showed that the representative G2d-0 virus strain from the 2024 season exhibited high lethality, and its virulence was also comparable to that of G2d-0 strains detected in previous seasons.
Amino acid sequence analysis of virus strains isolated up to November 2024 indicated no resistance to common antiviral drugs and no mutations associated with increased transmission in mammals. Therefore, the immediate risk of these viruses causing an epidemic among humans is considered low.
Migratory birds wintering in Japan are believed to harbor the virus until they return to their breeding grounds in early spring, potentially spreading it further as they migrate across Japan. Given this risk, stringent biosecurity measures are essential to prevent virus entry into poultry facilities. Additionally, early detection efforts must be strengthened to minimize outbreaks.

Funding
Budget: "Development of Livestock Infectious Disease Control Technology to Realize Livestock Industry Resilient to Emergence of New Infectious Diseases" (JPJ008617.23812859), a part of "Commissioned Research for Promotion of Comprehensive Regulatory Science for Stable Supply of Safe Agricultural, Livestock and Marine Products" of the Ministry of Agriculture, Forestry and Fisheries of Japan.
| For Inquiries |




